R workshop tutorial
Warning
Since the HPCC OS transition to Ubuntu in summer 2024, R modules have changed. This tutorial applies to R in general; however, HPCC-specific module commands need to be updated by the users at the moment. We will revamp R-related tutorials in the near future.
Preparation
- Basic knowledge of R language, Linux, and the HPCC environment.
- Login
ssh YourAccount@hpcc.msu.edussh dev-amd20
- We will be using R 4.0.2. To load
it:
module purge; module load GCC/8.3.0 OpenMPI/3.1.4 R/4.0.2 - Copy files that we are using in this workshop to your home
directory:
cp -r /mnt/research/common-data/workshops/R-workshop .
R startup
When an R session starts, it looks for and loads two hidden configuration files, .Renviron and .Rprofile.
.Renviron: contains environment variables to be set in R sessions.Rprofile: contains R commands to be run in R sessions
The following search order is applied: your current working directory,
your home directory, and the system-wide R_HOME/etc/ (you can use R
command R.home() to check path of R_HOME). Below are examples of the
two files which have been placed in our R workshop directory. You need
to use ls -a to list them since they are "hidden" files.
.Rprofile (an example)
cat("Sourcing .Rprofile from the R-workshop directory.\n")
# To avoid setting the CRAN mirror each time you run install.packages
local({
options(repos = "https://repo.miserver.it.umich.edu/cran/")
})
options(width=100) # max number of columns when printing vectors/matrices/arrays
.First <- function() cat("##### R session begins for", paste(Sys.getenv("USER"),".", sep=""), "You are currently in", getwd(), "#####\n\n")
.Last <- function() cat("\n##### R session ends for", paste(Sys.getenv("USER"),".", sep=""), "You are currently in", getwd(), "#####\n\n")
.Renviron (an example)
R_DEFAULT_PACKAGES="datasets,utils,grDevices,graphics,stats,methods"
R_LIBS_USER=/mnt/home/longnany/Rlibs
Let's run a short Rscript command to see what we get:
$ Rscript -e 'date()'
Notes:
- Personalizing these two files can reduce code portability.
-
If you don't want R or Rscript to read any
.Renvironor.Rprofilefiles when starting an R session, use option--vanilla.A caveat: if you explicitly export an R environment variable, such as
export R_LIBS_USER=~/Rlibs, then adding--vanillawill not ignore its value. See below the result.$ Rscript --vanilla -e '.libPaths()' # .libPaths() is used to see the directories where R searches for libraries [1] "/opt/software/R/4.0.2-foss-2019b/lib64/R/library" $ export R_LIBS_USER=~/Rlibs $ Rscript --vanilla -e '.libPaths()' [1] "/mnt/home/longnany/Rlibs" [2] "/opt/software/R/4.0.2-foss-2019b/lib64/R/library"
Rscript one-liner
The general format of Rscript:
Rscript [options] [-e expression1 -e expression2 ... | source file] [args]
-
options: can be multiple; all beginning with--(e.g.,--vanillaas mentioned above). To learn about all the options, runRscript --helpon the command line. -
expression1, expression2 ...: can be one or multiple. They are R commands. -
source file: R source code. -
args: arguments to be passed.
You may have both expressions and source file present in your Rscript line. Here are a few one-liner examples:
Rscript one-liner examples
# Ex1: simple loop
$ Rscript -e 'for (i in 1:5) print(paste("g", i))'
# Ex2: print time
$ Rscript -e 'date()'
# Ex3: quick math (calculating quotient and remainder)
$ Rscript -e '128 %/% 11' -e '128 %% 11'
# Ex4: get help for command "paste"
$ Rscript -e 'help(paste)'
# Ex5: used in conjunction with pipe.
# Generate three sets of random Normal variables with different means (sd all being one); means are given in file test.dat.
$ cat > test.dat # ctrl+D to go back when done typing
1
10
20
$ cat test.dat | Rscript -e 'input_con <- file("stdin"); open(input_con); while (length(oneLine <- readLines(con = input_con, n = 1, warn = FALSE)) > 0) {print(rnorm(5,mean=as.numeric(oneLine)))};close(input_con)'
Using Rscript with source code
a. simple usage:
Instead of using '-e your_commands', we now put R commands in a source file and run it with Rscript. Below is an R script file and we can run Rscript multiplots.R to get a PDF file multiplots.pdf. To view it, run evince multiplots.pdf. Or, you can go to our OnDemand File Browser to open the file in your web browser.
multiplots.R: a very simple example of making 4 plots on the same page
pdf("multiplots.pdf")
par(mfrow=c(2,2))
for (i in 1:4) plot(1:10, 1:10, type="b", xlab=bquote(X[.(i)]), ylab=bquote(Y[.(i)]), main=bquote("Multiple plots: #" ~ .(i)))
dev.off()
b. passing command line arguments:
We can also pass arguments to our R script, as shown in the example below.
args-1.R
args <- commandArgs(trailingOnly = TRUE)
nargs <- length(args)
for (i in 1:nargs) {
cat("Arg", i, ":", args[i], "\n")
}
cat("Generating",as.numeric(args[nargs-1]), "normal variables with mean =", as.numeric(args[nargs]), "and sd = 1 \n")
rnorm(as.numeric(args[nargs-1]), mean=as.numeric(args[nargs]))
Running script args-1.R:
$ Rscript args-1.R 5 3
Sourcing .Rprofile from the 11092017-R-Workshop directory.
##### R session begins for longnany. You are currently in /mnt/home/longnany/Documents/11092017-R-Workshop #####
Arg 1 : 5
Arg 2 : 3
Generating 5 normal variables with mean = 3 and sd = 1
[1] 2.707162 3.677923 3.192272 2.531973 3.699060
##### R session ends for longnany. You are currently in /mnt/home/longnany/Documents/11092017-R-Workshop #####
c. processing command line arguments with {getopt}:
args-2.R
require("getopt", quietly=TRUE)
spec = matrix(c(
"Number", "n", 1, "integer",
"Mean", "m", 1, "double"
), byrow=TRUE, ncol=4) # cf. https://cran.r-project.org/web/packages/getopt/getopt.pdf
opt = getopt(spec);
if (is.null(opt$Number)) {
n <- 5
} else {
n <- opt$Number
}
if (is.null(opt$Mean)) {
m <- 3
} else {
m <- opt$Mean
}
cat("Generating", n, "normal variables with mean =", m, "and sd = 1 \n")
rnorm(n=n, mean=m)
Running the script args-2.R:
# Use long flag names
$ Rscript --vanilla args-2.R --Number 10 --Mean -2
Generating 10 normal variables with mean = -2 and sd = 1
[1] -0.4776278 -1.7759145 -0.9977682 -2.6452126 -3.4050587 -2.2358362
[7] -1.2696362 -1.6213633 -2.7013074 -1.9271954
# Use short flag names
$ Rscript --vanilla args-2.R -n 10 -m -2
Generating 10 normal variables with mean = -2 and sd = 1
[1] -2.2241837 -1.6704711 0.1481244 0.2072124 -1.0385386 -1.5194874
[7] -2.6744478 -2.4683039 -0.7962113 -1.1901021
# No arguments provided so defaults are used
$ Rscript --vanilla args-2.R
Generating 5 normal variables with mean = 3 and sd = 1
[1] 3.951492 4.255879 4.485044 2.727223 3.039532
Submitting parallel jobs to the cluster using doParallel: single node, multiple cores
To submit a single-node job, we recommend the package doParallel.
The doParallel package is a "parallel backend" for the foreach package, making it possible to execute foreach loops in parallel.
Note
doParallel supports two functionalities: multicore and snow. The most important difference between them is that multicore can only run tasks on a single node (computer) whereas snow can execute tasks on different nodes in a cluster. This leads to different commands for registering the parallel backend. In our example here, we are interested in using the multicore-like parallelism, since we are trying to use the many cores on a single compute node in the cluster. To learn more about their differences, you can refer to this discussion.
Example R code R_doParallel_singlenode.R: run 10,000 bootstrap iterations of fitting a logistic regression model
library(doParallel)
# Registering a parallel backend, using the "multicore" functionality
registerDoParallel(cores=as.numeric(Sys.getenv("SLURM_CPUS_ON_NODE")[1]))
x <- iris[which(iris[,5] != "setosa"), c(1,5)]
trials <- 10000
ptime <- system.time({
r <- foreach(icount(trials), .combine=cbind) %dopar% {
ind <- sample(100, 100, replace=TRUE)
result1 <- glm(x[ind,2]~x[ind,1], family=binomial(logit))
coefficients(result1)
}
})[3]
cat("Time elapsed:", ptime, "\n")
cat("Currently registered backend:", getDoParName(), "\n")
cat("Number of workers used:", getDoParWorkers(), "\n")
print(str(r)) # column-binded result
Now, submit the job to the HPCC through the following SLURM script submit-doParallel.sbatch:
#!/bin/bash
# Job name:
#SBATCH --job-name=doParallel_test
#
# Number of nodes needed:
#SBATCH --nodes=1
#
# Tasks per node:
#SBATCH --ntasks-per-node=1
#
# Processors per task:
#SBATCH --cpus-per-task=4
#
# Memory per node:
#SBATCH --mem=500M
#
# Wall time (e.g. "minutes", "hours:minutes:seconds", "days-hours", "days-hours:minutes"):
#SBATCH --time=3:00:00
#
# Standard out and error:
#SBATCH --output=%x-%j.SLURMout
echo "This script is from ICER's R doParallel example"
module purge
module load GCC/8.3.0 OpenMPI/3.1.4 R/4.0.2
Rscript --vanilla R_doParallel_singlenode.R > R_doParallel_singlenode.Rout
Submission command: sbatch --constraint="[intel18|amd20|amd22]" submit-doParallel.sbatch
The output file R_doParallel_singlenode.Rout should look like:
Time elapsed: 8.946
Currently registered backend: doParallelMC
Number of workers used: 4
num [1:2, 1:10000] -14.6 2.26 -11.86 1.91 -7.75 ...
- attr(*, "dimnames")=List of 2
..$ : chr [1:2] "(Intercept)" "x[ind, 1]"
..$ : chr [1:10000] "result.1" "result.2" "result.3" "result.4" ...
NULL
Submitting parallel jobs to the cluster using pbdMPI: multiple nodes
MPI stands for Message Passing Interface, and is the standard for managing multi-node communication. The R package
Rmpi is an implementation of it for R. Because of its complicated usage, here we will use another R package, pbdMPI, which greatly simplifies the use of MPI from R.
pbdMPI is a more recent R MPI package developed under the pbdR project, which simplifies MPI interaction and thus reduces the traffics of node-to-node communication. It works in Single Program/Multiple Data (SPMD) mode, which is an important distinction as compared with Rmpi.
As an illustration, we consider the problem of computing the log likelihood of data following a 2-dimensional Multi-Variate Normal (MVN) distribution. The example below applies Cholesky decomposition on the 2-by-2 covariance matrix (code line 25 below), solves a system of linear equations (line 30), followed by some matrix/vector operation (line 31). Line 30 and 31 are where MPI plays a vital role via distributed computing.
Below is a graphic illustration of solving a system of linear equations by part so as to be able to distribute the task. To learn more about the underlying mechanism, you can take a look at this note.
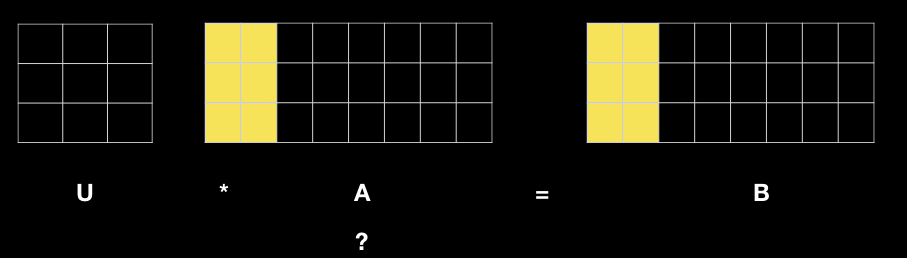
MVN.R: MPI in SPMD mode
# Load pbdMPI and initialize the communicator
library(pbdMPI, quiet = TRUE)
init()
# Check processes
comm.cat("All processes start...\n\n")
msg <- sprintf("I am rank %d on host %s of %d processes\n", comm.rank(), Sys.info()["nodename"], comm.size())
comm.cat(msg, all.rank=TRUE, quiet=TRUE) # quiet=T tells each rank not to "announce" itself when it's printing
set.seed(1234)
N <- 100
p <- 2
X <- matrix(rnorm(N * p), ncol = p)
mu <- c(0.1, 0.2)
Sigma <- matrix(c(0.9, 0.1, 0.1, 0.9), ncol = p)
# Load data partially by processors
id.get <- get.jid(N)
X.spmd <- matrix(X[id.get, ], ncol = p)
comm.cat("\nPrint out the matrix on each process/rank:\n\n", quiet=TRUE)
comm.print(X.spmd, all.rank=TRUE, quiet=TRUE)
# Cholesky decomposition
U <- chol(Sigma) # U'U = Sigma
logdet <- sum(log(abs(diag(U))))
# Call R's backsolve function for each chunk of the data matrix X (i.e. B.spmd)
B.spmd <- t(X.spmd) - mu
A.spmd <- backsolve(U, B.spmd, upper.tri = TRUE, transpose = TRUE) # U'A = B
distval.spmd <- colSums(A.spmd * A.spmd)
# Use sum as the reduction operation
sum.distval <- allreduce(sum(distval.spmd), op = "sum")
total.logL <- -0.5 * (N * (p * log(2 * pi) + logdet * 2) + sum.distval)
# Output
comm.cat("\nFinal log-likelihood:\n\n", quiet=TRUE)
comm.print(total.logL, quiet=TRUE)
finalize()
SLURM submission script: submit-pbdMPI.sbatch:
#!/bin/bash
# Job name:
#SBATCH --job-name=pbdMPI_test
#
# Number of MPI tasks:
#SBATCH --ntasks=20
#
# Processors per task:
#SBATCH --cpus-per-task=1
#
# Memory:
#SBATCH --mem-per-cpu=800M
#
# Wall clock limit (e.g. "minutes", "hours:minutes:seconds", "days-hours", "days-hours:minutes"):
#SBATCH --time=30
#
# Standard out and error:
#SBATCH --output=%x-%j.SLURMout
echo "This script is from ICER's R pdbMPI example"
echo "SLURM_NTASKS: $SLURM_NTASKS"
module purge
module load GCC/8.3.0 OpenMPI/3.1.4 R/4.0.2
# Suppress warnings about forks and missing CUDA libraries
export OMPI_MCA_mpi_warn_on_fork=0
export OMPI_MCA_mpi_cuda_support=0
mpirun -n $SLURM_NTASKS Rscript --vanilla MVN.R > MVN.Rout
Now, submit the job by sbatch --constraint="[intel18|amd20|amd22]" submit-pbdMPI.sbatch. When finished, MVN.Rout should contain the following information:
COMM.RANK = 0
All processes start...
I am rank 0 on host lac-419 of 20 processes
I am rank 1 on host lac-420 of 20 processes
... ...
I am rank 18 on host lac-421 of 20 processes
I am rank 19 on host lac-421 of 20 processes
Print out the matrix on each process/rank:
[,1] [,2]
[1,] -0.1850210 -0.2771503
[2,] -0.8596753 -0.1081122
[3,] -1.0927853 0.8690750
[4,] -0.5831948 0.8032846
[5,] 1.0796870 0.2354514
[,1] [,2]
[1,] 0.05274001 -0.8410549
[2,] -0.92568393 0.9461704
[3,] 1.01632804 0.6875080
[4,] 0.34271061 0.4905065
[5,] 0.84192956 -1.6685933
... ...
[,1] [,2]
[1,] -0.55673659 -0.1229023
[2,] -0.03156318 -0.8501178
[3,] -1.28832627 -1.3115801
[4,] -0.47546826 -0.4856559
[5,] 0.81134183 0.7499220
[,1] [,2]
[1,] -0.95620195 0.6560605
[2,] 0.04671396 0.6093924
[3,] 0.18986742 0.2077641
[4,] 0.73281327 -1.0452661
[5,] 2.27968859 1.0611428
Final log-likelihood:
[1] -283.2159
You can download more R pbdMPI examples from here.